- Dissolving Charged Peptides
- Dissolving Peptides
- Dissolving Uncharged Peptides
- Lyophilized Peptides Storage
- Peptide Solubility
- Peptide Stability and Degradation
A peptide is acidic if the overall net charge of the peptide is negative. For an acidic peptide, if the total
number of charges of the peptide at pH 7 is greater than 25% of the total number of residues, add a small amount
of 0.1M ammonium bicarbonate to dissolve the peptide and dilute it with water to the desired concentration.
Make certain that the resulting pH of the peptide solution is about 7 and adjust the pH as needed.
A peptide is basic if the overall net charge of the peptide is positive. For a basic peptide, if the total
number of charges of the peptide at pH 7 is between 10-25% of the total number of residues, add a small amount
of 25% acetic acid to dissolve the peptide and dilute it with water to the desired concentration.
A peptide is considered neutral if the overall net charge of the peptide is zero. If the total number of
charges is greater than 25% of the total number of residues, use the strategy described in section 1. If the
total number of charges is between 10-25% of the total number of residues, use organic solvents as recommended
elsewhere in this document.
If the total number of charges of a peptide is less than 10% of the total number of residues, use of organic
solvents is recommended.
Dissolve peptides in 0.1% aqueous acetic acid to yield a target concentration of 1-5 mg/mL. Use sonication if necessary.
Small amounts of dilute(10%) aqueous acetic acid for basic peptide antigen or aqueous ammonia for acidic peptides may
help dissolution of these peptides.
If peptides are still insoluble, add acetonitrile up to 20%(v/v), and use sonication to help dissolution.
Lyophilize any remaining insoluble peptides to remove water, acetic acid and acetonitrile. When the peptides are
completely dry, add neat DMF or DMSO (dropwise) until the peptides are dissolved. Slowly dilute the solution with
water to desired concentration. If precipitation occurs during dilution, add more DMF or DMSO (dropwise) to dissolve
the precipitate.
A peptide is acidic if the overall net charge of the peptide is negative. For an acidic peptide, if the total number
of charges of the peptide at pH 7 is greater than 25% of the total number of residues, add a small amount of 0.1M
ammonium bicarbonate to dissolve the peptide and dilute it with water to the desired concentration. Make certain
that the resulting pH of the peptide solution is about 7 and adjust the pH as needed.
A peptide is basic if the overall net charge of the peptide is positive. For a basic peptide, if the total number of
charges of the peptide at pH 7 is between 10-25% of the total number of residues, add a small amount of 25% acetic acid
to dissolve the peptide and dilute it with water to the desired concentration.
A peptide is considered neutral if the overall net charge of the peptide is zero. If the total number of charges is greater
than 25% of the total number of residues, use the strategy described in section 1. If the total number of charges is between
10-25% of the total number of residues, use organic solvents as recommended elsewhere in this document.
If the total number of charges of a peptide is less than 10% of the total number of residues, use of organic solvents is recommended.
Lyophilized peptides are stable at room temperature for weeks. Upon arrival, always store lyophilized peptides in a freezer at
– 20°C for maximum stability. For short-term storage, it is recommended to store peptides at 4°C.
Lyophilized peptides are often hydroscopic. For best results, maintain product for synthetic peptide in a dry environment. When
preparing peptides for use or peptide antigen design for example, allow to equilibrate to room temperature before opening the
container, reseal the vial quickly after weighing out desired quantity.
Assign a value of -1 to each acidic residue (D, E, and C-terminal COOH)
Assign a value of +1 to each basic residue (K, R and the N-terminal NH2)
Assign a value of +1 to each H residue at pH<6 and zero at pH >6.
Count the total number of charges of the peptide at pH 7 (all D, E, K, R, C-terminal COOH, and C-terminal NH2).
Calculate the overall net charge of the peptide
Hydrolysis
Peptides containing Asp (D)
Sequence contains Asp-Pro (D-P)
Similarly, if Asp-Gly (D-G) is present in the sequence
Sequences containing Ser (S)
Peptides containing Asp (D)
Sequence contains Asp-Pro (D-P)
Similarly, if Asp-Gly (D-G) is present in the sequence
Sequences containing Ser (S)
Deamidation sequences containing:
Asn-Gly (N-G)
Gln-Gly (Q-G)
Asp-Gly (D-G)
Asn-Gly (N-G)
Gln-Gly (Q-G)
Asp-Gly (D-G)
Oxidation
The Cys (C) and Met (M)
The Cys (C) and Met (M)
Diketopiperazine and pyroglutamic acid formation
Gly (G) is in the third position from the N-terminus
Pro (P) or Gly (G) is in position 1 or 2
Gly (G) is in the third position from the N-terminus
Pro (P) or Gly (G) is in position 1 or 2
- Antibody Modifications
- Fluorescent Modifications
- FRET Peptides
- Peptide Cyclization
- Standard Modifications
KLH, BSA, OVA Conjugates
Peptide-protein conjugates are used for custom antibody production against peptides. Peptides alone are mostly too small to
elicit a sufficient immune response, so carrier proteins containing many epitopes help to stimulate T-helper cells, which
help induce the B-cell response. It is important to remember that the immune system reacts to the peptide-protein conjugate
as a whole so there will always be a portion of antibodies to the peptide synthesis, the linker and the carrier protein.
Among the most common carrier proteins one can find:
KLH (Keyhole Limpet Hemocyanin), a copper containing, non-heme protein
found in arthropods and mollusca. It is isolated from Megathura crenulata and has a MW of 4.5 x 105 ~ 1.3 x 107 Da.
KLH is the most commonly selected carrier due to its higher immunogenicity compared to BSA.
BSA (Bovine Serum Albumin), a plasma protein in cattle, belonging to
the most stable and soluble albumins. It has a MW of 67 x 103 Da containing 59 lysines. About 30-35 of these primary amines
are accessible for linker conjugation, which makes BSA a popular carrier protein for weak antigenic compounds. A
disadvantage of BSA is that it is used in many experiments as a blocking buffer reagent. If antisera against peptide-BSA
conjugates are used in such assays, false positives can occur, because these sera also contain antibodies to BSA.
OVA (Ovalbumin), a protein isolated from hen egg whites, with a MW
of 45 x 103 Da. It is a good choice as second carrier protein to verify if antibodies are specific for the peptide alone
and not the carrier protein (e.g. BSA).
MAPs (Multiple peptide antigen)
MAPs are branched peptides that can be used for direct immunization to produce antibodies. MAPs are usually big enough to
raise the immune response.
The antigenic peptide of interest is being synthesized directly on the branched MAP structure. MAPs are available as MAP 4
(4 branches) or MAP 8 (8 branches) molecules:
Schematic graph of a MAP 8 and a peptide-protein conjugate:
AMC (7-Amino-4-methyl-coumarin)
UV-excitable dye, used in enzyme assays using cuvettes or flow cytometry.
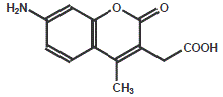
| Dye | Excitation maximum | Emission maximum | Molar Extinction Coefficient |
| AMC | 353 nm | 422 nm | 19.000 |
Cy3, Cy5
Cy3, Cy5 are dyes with extremely high extinction coefficients and fluorescence. Thus, they are especially suitable for very
sensitive localization assays of peptides in cells. Their disadvantage is the high instability of the molecules under peptide
synthesis conditions. Therefore the yields are comparatively low.
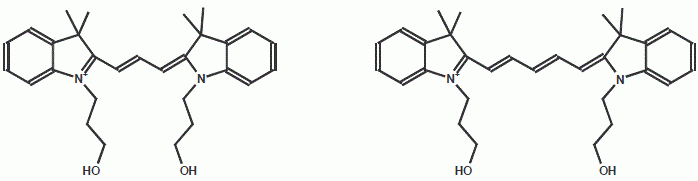
| Dye | Excitation maximum | Emission maximum | Molar Extinction Coefficient |
| Cy™3 | 550 nm | 570 nm | 150.000 |
| Cy™5 | 650 nm | 670 nm | 250.000 |
Dabcyl
Dabcyl is a non-fluorescent dye predominantly used as a quencher for other fluorophores (esp. Fluorescent type dyes, EDANS.).
If Dabcyl is coupled to a peptide in close proximity to a fluorophore, it absorbs the emitted light of the fluorophore.
Enlarging this distance (i.e. by enzymatic cleavage of the peptide) results in excitation of the fluorophore with an
emission signal that can be detected.
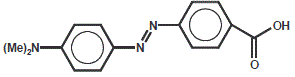
| Dye | Excitation maximum | Emission maximum | Molar Extinction Coefficient |
| Dabcyl | 453 nm | none | 32.000 |
Dansyl
Dansyl is also used as a fluorophore quencher. Unlike Dabcyl, it inherits own fluorescence and thus might not be as useful
for highly sensitive assays
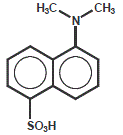
| Dye | Excitation maximum | Emission maximum | Molar Extinction Coefficient |
| Dansyl | 335 nm | 526 nm | 4.600 |
2,4-Dinitrophenyl (DNP)
2,4-Dinitropheny is a non-fluorescent dye that can be used as a fluorophore quencher (see Dabcyl for more details).
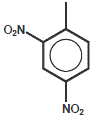
| Dye | Excitation maximum | Emission maximum | Molar Extinction Coefficient |
| DNP | 348 nm | none | 18.000 |
DNP-Lysine
DNP-Lysineis a non-fluorescent dye that can be used as a fluorophore quencher, for custom peptide (see Dabcyl for more details).
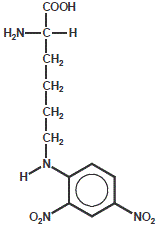
| Dye | Excitation maximum | Emission maximum | Molar Extinction Coefficient |
| DNP-Lysine | 348 nm | none | 18.000 |
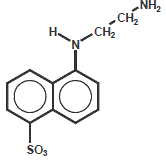
EDANS (5-((2-aminoethyl)amino)napthalene-1-sulfonic acid)
EDANS is a commonly used dye in FRET (fluorescence resonance energy transfer) peptides in combination with Dabcyl as quencher.
| Dye | Excitation maximum | Emission maximum | Molar Extinction Coefficient |
| EDANS | 335 nm | 493 nm | 5.900 |
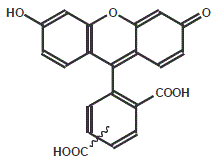
Fluorescent
Fluorescent is the commonly used fluorescent dye in confocal laser-scanning microscopy and flow cytometry applications.
| Dye | Excitation maximum | Emission maximum | Molar Extinction Coefficient |
| Fluorescent | 495 nm | 520 nm | 83.000 |
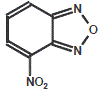
NBD (7-nitrobenz-2-oxa-1, 3-diazole)
NBD is a fluorescent dye, used for amine modification.
| Dye | Excitation maximum | Emission maximum | Molar Extinction Coefficient |
| NBD | 486 nm | 543 nm | 27.000 |
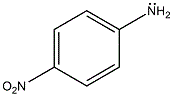
p-Nitro-Aniline
p-Nitro-Aniline is a chromogen used as colorimetric enzyme substrate in many standard enzyme assays in cuvettes.
Rhodamine B
Rhodamine B represents one among a numerous range of rhodamine dyes, used in fluorescent peptide assays.
| Dye | Excitation maximum | Emission maximum | Molar Extinction Coefficient |
| Rhodamine | 550 nm | 580 nm | 90.000 |
Tamra
Tamra is the most commonly used rhodamine dye in fluorescence assays.
| Dye | Excitation maximum | Emission maximum | Molar Extinction Coefficient |
| Tamra | 544 nm | 576 nm | 90.000 |
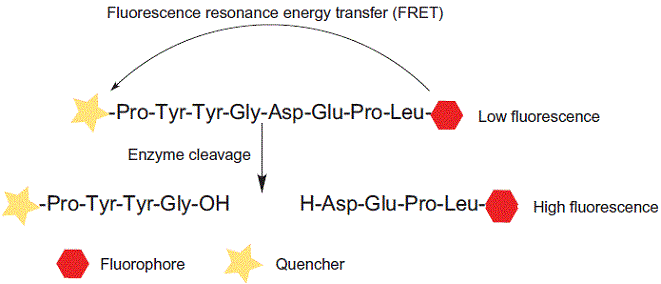
FRET stands for fluorescence resonance energy transfer, or Förster resonance energy transfer. FRET is a mechanism describing
excited state energy transfer from the initially excited donor to an acceptor. The donor molecules typically emit at shorter
wavelengths that overlap with the absorption of acceptors. The process is a distance-dependent interaction between the
electronic excited states of two molecules without emission of a photon.
FRET can be used for conformational investigation of peptide folding. FRET peptides are widely used as suitable substrates in
enzyme studies, such as:
Functional characterization of peptidases / proteases / kinases / phosphatases
Kinetic characterization of peptidases / proteases / kinases / phosphatases
Screening and detection of new proteolytic enzyme
Kinetic characterization of peptidases / proteases / kinases / phosphatases
Screening and detection of new proteolytic enzyme
Chromophore/Fluorophore – Quencher Pairs
| Chromophore/Fluorophore | Lambda max (nm) | Lambda em (nm) | Quencher |
| MAC | 432 | 441 | - |
| Dabcyl | 453 | - | - |
| Dansyl | 335 | 526 | Dabcyl |
| Dnp | 348 | - | - |
| EDANS | 341 | 471 | Dabcyl |
| Mca | 328 | 393 | Dnp |
| FAM | 494 | 518 | Dabcyl |
| TAMRA | 555 | 580 | - |
| Mca | Dpa | - | - |
How does a quencher work?
A quencher is very efficient at absorbing certain wavelengths. When near a dye that emits at the absorbed wavelength, the
light is “quenched”, and no longer visible. Quenchers are very similar in structure to dyes. The difference is that they
emit in undetectable ranges, or in undetectable amounts. The ability to quench is a function of distance from the dye in
most cases. Molecular beacons are effective in that the quencher actually comes in contact with the dye. Different quenchers
are best for different dyes.
Can a dye be conjugated to protein or peptide internally?
Yes. Use the succinimidyl ester version of the dye and conjugate it to either a N-terminal and Lysine side-chain amino linker.
Other options include using a Lys-FAM or Lys-TAMRA for Solid Phase Peptide Synthesis. These dyes are already conjugated to
the Lys side-chain amino linker.
What is the difference between a 5-FAM and FITC labeled protein and peptide?
Both 5-FAM (5-carboxyfluorescein, single isomer) and FITC (fluorescein-5-isothiocyanate, isomers) are forms of the fluorescent
dye fluorescein. FITC refers to a particular form of reactive species, the isothiocyanate, of the dye. It yields a urea linkage
upon reaction with a primary amine. 5-FAM is the preferred reagent for labeling a protein or peptide. It results in an amide
bond when reacted with a primary amine. The chemistry is more robust and better yielding. Furthermore, it has been shown that
5-FAM is less susceptible to photo-bleaching. The 5-FAM and FITC conjugated protein and peptide have similar spectral properties.
Can I use a dye or modification that is not listed here?
Yes. Tell us the dye or modification you are looking to use; we will find out if it is commercially available. If it is not,
there is a possibility we can make it in-house. If neither of these is an option we will do our best to see if there is
another dye or modification that can give you similar results. If any of these options work for you, we will then send you
a quotation.
What are the absorption max and emission max values for my fluorescent labeled protein or peptide?
Spectra data can be found (click here). These values are provided by the manufacturer of the fluorescent dye and are generally
calculated from the free dye not attached to a protein or peptide. As a general rule, these values work fine for the common
protein or peptide user as there is little change, if any at all, when the dye is attached to a protein or peptide.
The particular base composition of a protein or peptide can play a role as can pH in many cases, particular fluorescein and
related dyes.
Can you make a peptide with two different dyes or other conjugate?
The answer is complicated in that there are many ways to place different conjugations on a peptide, but we are still limited
by what is available to us for conjugation and compatibility issues. The easiest way is if one or both of the conjugates are
available as thiol of cysteine and/or support bound reagents. If at least one reagent is available, the other conjugate can
often be attached through a primary amine at the other end of the peptide. Chemical compatibility is avoided in this case.
If neither conjugate is available as a thiol of cysteine or support bound reagent, then we often will use an orthogonal
linker scheme incorporating both a thiol linker and an amino linker. This requires that one of the conjugates is available
as a maleimide, which is thiol specific under neutral pH conditions. Most of all the commonly used dyes are available in
both the succinimidyl and maleimidyl ester forms. The most common, such as fluorescein, TET, HEX, TAMRA, Cy™5 and Cy3 are
also available as primary amine or support bound reagents. We will help you design your specific peptide when you place your
order. Also, see our Products section on dyes for more information on what is available.
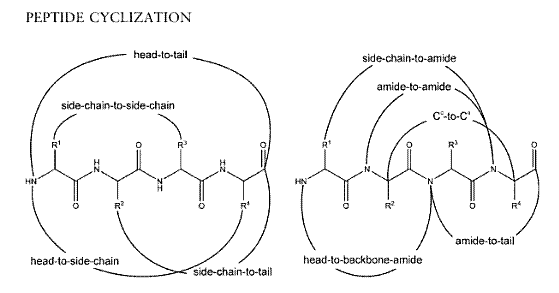
Different modes of cyclization for peptides:
Left: Conventional cyclization
Right: Backbone cyclization with all possible connections to backbone amides
Right: Backbone cyclization with all possible connections to backbone amides
Biotin and Desthiobiotin
Biotin (or vitamin H) is a small biologically active molecule with a molecular weight of 244.31 Da. It acts as a co-enzyme in
living cells. With its highly specific affinity towards streptavidin, it is used in various biotechnology assays &peptide
synthesis for quality and quantity testing.
Desthiobiotin: Binds to streptavidin but can be displaced by biotin. Useful when you need to get your peptide out of a binding
experiment. THE molecular weight: 214.31 Da.
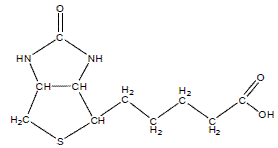
Farnesyl
Farnesyl is a potential substrate to study demethylase activity in enzyme assays.
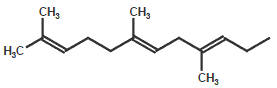
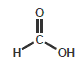
Formic acid (Formyl)
Myristic acid (Myristoyl)

Palmitic acid (Palmitoyl)

Stearic acid (Stearyl)

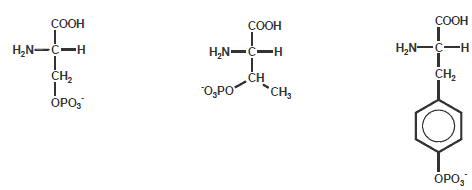
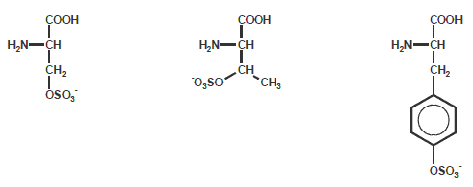
Phosphorylation Peptide Modification
Phosphorylationof Ser, Thr and Tyr is one of the more common modifications of amino acids in nature. Many hormones can adapt
the activity of specific enzymes by increasing their phosphorylation state of Ser or Thr residues. Growth factors (like insulin)
can trigger phosphorylation of Tyr.
The phosphate groups on these amino acids can be quickly removed, thus Ser, Thr and Tyr function as molecular switches during
regulation of cellular processes (e.g. cancer proliferation).
Succinic acid (Succinyl)
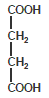
Sulfurylation
Sulfurylation at Ser, Thr and Tyr is another Peptide Modification of amino acids in nature. Activity of many enzymes depends on
the oxidation state of SH-groups in these residues.
| 2nd base | |||||
| U | C | A | G | ||
| 1st base | U |
UUU (Phe/F)Phenylalanine UUC (Phe/F)Phenylalanine UUA (Leu/L)Leucine UUG (Leu/L)Leucine |
UCU (Ser/S)Serine UCC (Ser/S)Serine UCA (Ser/S)Serine UCG (Ser/S)Serine |
UAU (Tyr/Y)Tyrosine UAC (Tyr/Y)Tyrosine UAA Ochre (Stop) UAG Amber (Stop) |
UGU (Cys/C)Cysteine UGC (Cys/C)Cysteine UGA Opal (Stop) UGG (Trp/W)Tryptophan |
| C |
CUU (Leu/L)Leucine CUC (Leu/L)Leucine CUA (Leu/L)Leucine CUG (Leu/L)Leucine |
CCU (Pro/P)Proline CCC (Pro/P)Proline CCA (Pro/P)Proline CCG (Pro/P)Proline |
CAU (His/H)Histidine CAC (His/H)Histidine CAA (Gln/Q)Glutamine CAG (Gln/Q)Glutamine |
CGU (Arg/R)Arginine CGC (Arg/R)Arginine CGA (Arg/R)Arginine CGG (Arg/R)Arginine |
|
| A |
AUU (Ile/I)Isoleucine AUC (Ile/I)Isoleucine AUA (Ile/I)Isoleucine AUG (Met/M)Methionine, (Start) |
ACU (Thr/T)Threonine ACC (Thr/T)Threonine ACA (Thr/T)Threonine ACG (Thr/T)Threonine |
AAU (Asn/N) Asparagine AAC (Asn/N)Asparagine AAA (Lys/K)Lysine AAG (Lys/K)Lysine |
AGU (Ser/S) Serine AGC (Ser/S)Serine AGA (Arg/R) Arginine AGG (Arg/R)Arginine |
|
| G |
GUU (Val/V)Valine GUC (Val/V)Valine GUA (Val/V)Valine GUG (Val/V)Valine |
GCU (Ala/A)Alanine GCC (Ala/A)Alanine GCA (Ala/A)Alanine GCG (Ala/A)Alanine |
GAU (Asp/D)Aspartic acid GAC (Asp/D)Aspartic acid GAA (Glu/E)Glutamic acid GAG (Glu/E)Glutamic acid |
GGU (Gly/G)Glycine GGC (Gly/G)Glycine GGA (Gly/G)Glycine GGG (Gly/G)Glycine |
|
Reverse Codon Table
This table shows the 20 standard amino acids used in proteins, and the codons that code for each amino acid.
| Ala | A | GCU, GCC, GCA, GCG | Leu | L | UUA, UUG, CUU, CUC, CUA, CUG |
| Arg | R | CGU, CGC, CGA, CGG, AGA, AGG | Lys | K | AAA, AAG |
| Asn | N | AAU, AAC | Met | M | AUG |
| Asp | D | GAU, GAC | Phe | F | UUU, UUC |
| Cys | QC | UGU, UGC | Pro | P | CCU, CCC, CCA, CCG |
| Gln | Q | CAA, CAG | Ser | S | UCU, UCC, UCA, UCG, AGU,AGC |
| Glu | E | GAA, GAG | Thr | T | ACU, ACC, ACA, ACG |
| Gly | G | GGU, GGC, GGA, GGG | Trp | W | UGG |
| His | H | CAU, CAC | Tyr | Y | UAU, UAC |
| Ile | I | AUU, AUC, AUA | Val | V | GUU, GUC, GUA, GUG |
| Start | AUG | Stop | UAG, UGA, UAA | ||
In general, a good peptide antigen has the following properties: protein surface location, flexible (usually loop) rather than helical
structure, complicate and unique sequence, not a post-translational modification site or functional site (unless the antibody is
intended to recognize such a site) and easy for synthesis. But the quantitative relationship between these properties and antigenic
strength is not clear
Peptide antigen design is usually done by two different approaches: predicting peptide’s physical-chemistry properties or making
prediction based on statistic results. Both approaches have their limitations. Physical chemistry properties such as peptide location
in a protein or its secondary structure, particular turn, are difficult to be predicted with high degree of accuracy. This is because
the problem themselves are parts of one of the most challenge areas of modern science —- protein folding. Another problem is that in
most cases, an isolated peptide in solution can not maintain its native conformation found in the protein. This is the major reason
that antigens designed based on known 3D structures are also often failed.
Although Cys residues are often added to peptides to enable crosslinking to a carrier protein, a peptide synthesized with many Cys
residues present can be difficult to handle and may not lead to a useful antibody. Multiple Cys residues may lead to the formation of
covalently linked aggregates. The Cys-rich regions of proteins may have some disulfide bonds. The linear peptide with reduced Cys would
therefore not represent the protein itself, which would be more structurally constrained.
Most peptide antigen requested range in length from 12 to 20 residues and are relatively easy to synthesize. No Cys residues should be
internal to the peptide sequence.
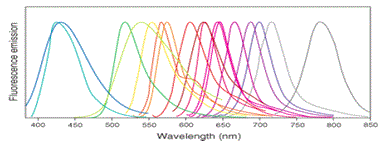
Fluorophores Absorption and Emission Data
The table lists the properties of the most commonly used fluorophore for peptide labeling of your interest in the life sciences. The
fluorophores absorbance wavelength from 325nm to 743nm (from blue color to red color), the emission wavelength from 386nm to 770nm.
| Fluorophore Dyes | Ex (nm) | Em (nm) |
| Hydroxycoumarin | 325 | 386 |
| Dansyl | 340 | 578 |
| 7-Amino-4-methylcoumarin | 351 | 430 |
| methoxycoumarin | 360 | 410 |
| Alexa fluor | 345 | 442 |
| aminocoumarin | 350 | 445 |
| MAC | 345 | 445 |
| Dabcyl | 453 | - |
| Cy2 | 490 | 510 |
| FAM | 495 | 517 |
| Alexa fluor 488 | 494 | 517 |
| Fluorescein FITC | 495 | 519 |
| Alexa fluor 430 | 430 | 545 |
| 5-Carboxyfluorescein (5-FAM) | 492 | 518 |
| Alexa fluor 532 | 530 | 555 |
| HEX | 535 | 556 |
| 5-TAMRA | 542 | 568 |
| Cy3 | 550 | 570 |
| TRITC | 547 | 572 |
| Alexa fluor 546 | 556 | 573 |
| Alexa fluor 555 | 556 | 573 |
| Dansyl | 340 | 578 |
| R-phycoerythrin (PE) | (489) | 565 |
| Rhodamine Red-X | 560 | 580 |
| Tamara | 565 | 580 |
| Cy3 | (512) | 550 |
| Cy3.5 | 581 | 596 |
| Rox | 575 | 602 |
| Alexa fluor 568 | 578 | 603 |
| Texas Red | 589 | 615 |
| Alexa fluor 594 | 590 | 617 |
| Alexa fluor | 621 | 639 |
| Alexa fluor 633 | 650 | 668 |
| Cy5 | (625) | 650 |
| Alexa fluor 660 | 663 | 690 |
| Cy5.5 | 675 | 694 |
| TruRed | 490; 675 | 695 |
| Alexa fluor 680 | 679 | 702 |
| Cy7 | 743 | 767 |


